publications
2023
-
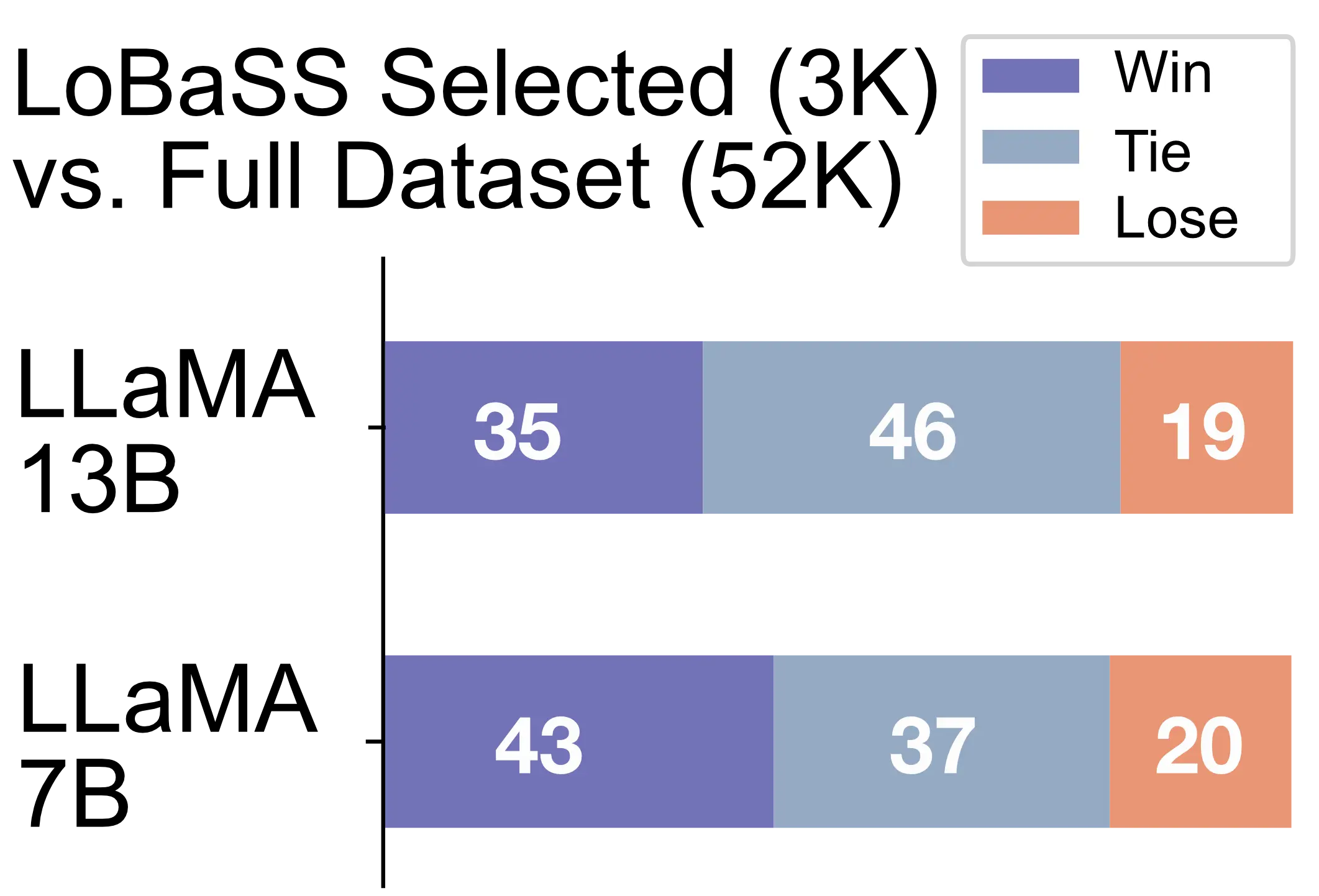 arXiv, Oct 2023
arXiv, Oct 2023We propose a loss-based LLM post-training data selection method method, which compresses Alpaca dataset by 16x while improving model performance on AlpacaEval.
-
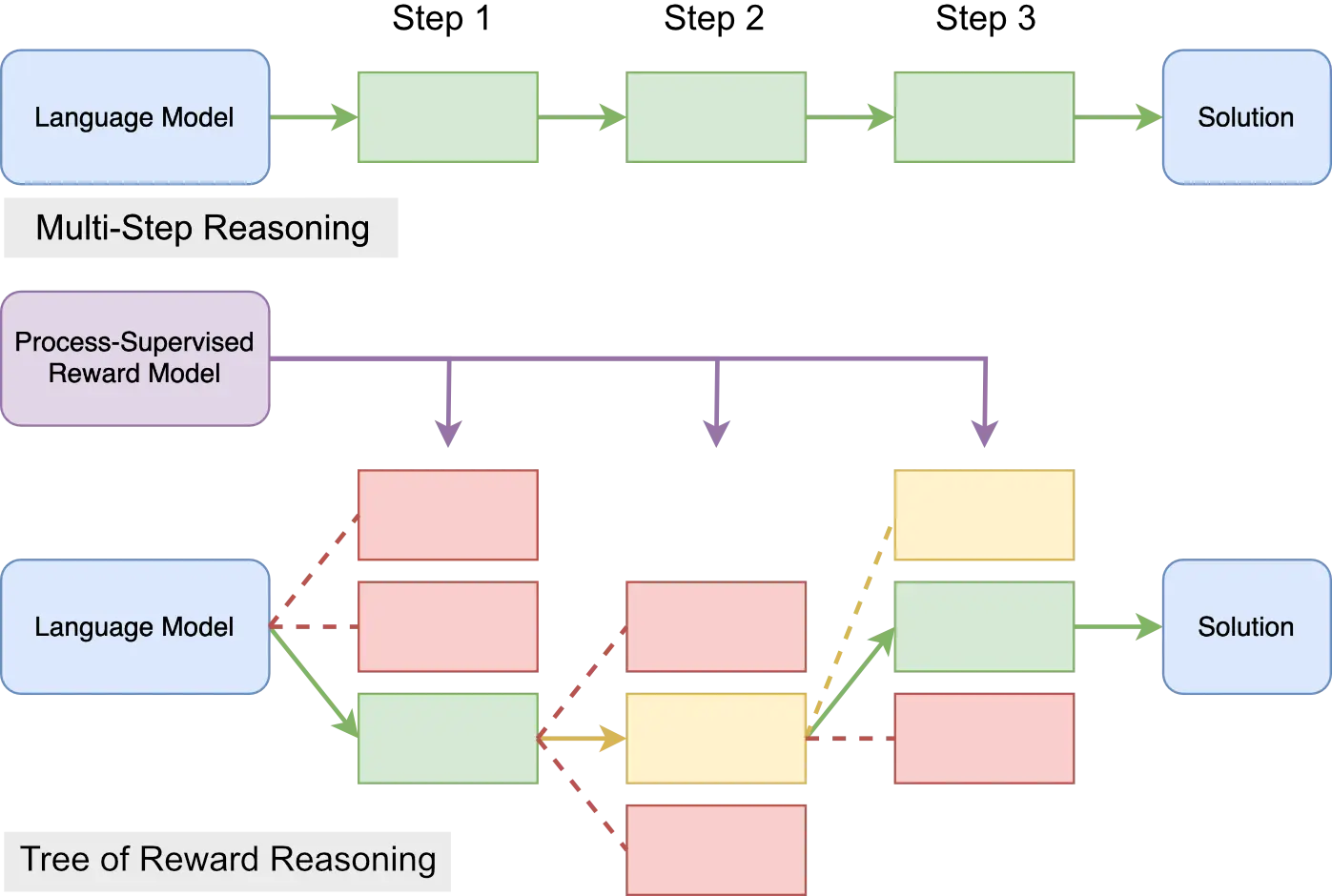 arXiv, Oct 2023
arXiv, Oct 2023We propose a heuristic greedy search algorithm based on step-level reward for improved LLM reasoning capabilities during inference. We also present a new automated way to generate massive amount of step-level reward signal for code generation tasks based on mutation testing.
-
 arXiv, Oct 2023
arXiv, Oct 2023We propose Video-Teller, a video-language foundation model with fine-grained modality alignment to enhance video-to-text generation tasks.
-
 arXiv, Oct 2023
arXiv, Oct 2023We introduce SimVLG, a one-staged single-loss framework based on Token Merging, for pre-training computationally intensive vision-language generative models using frozen pre-trained large language models.
-
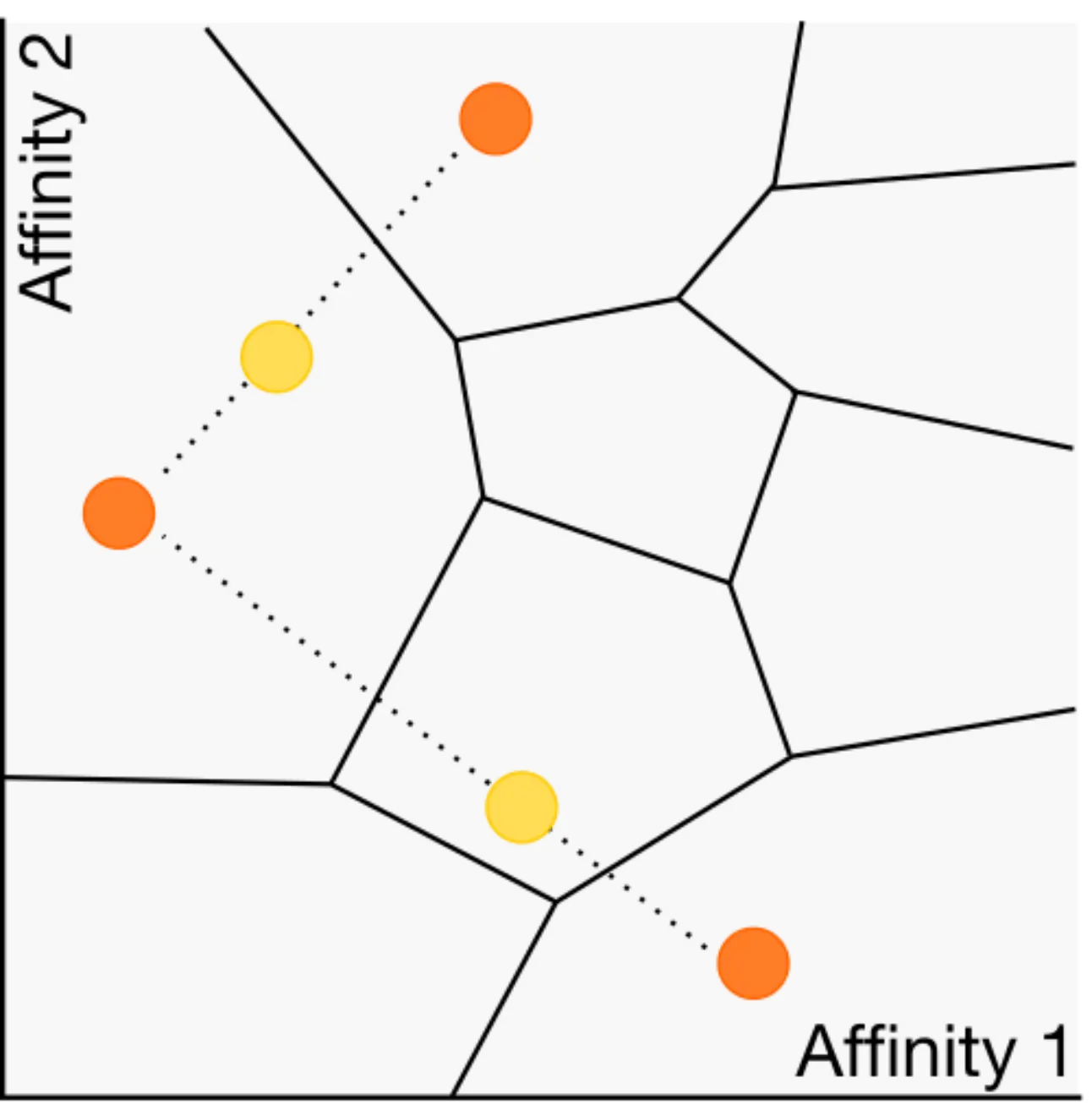 US Patent US11674937B2, , Jun 2023
US Patent US11674937B2, , Jun 2023We developed a biomimetic method and apparatus for encoding odorants, which has been patented.
-
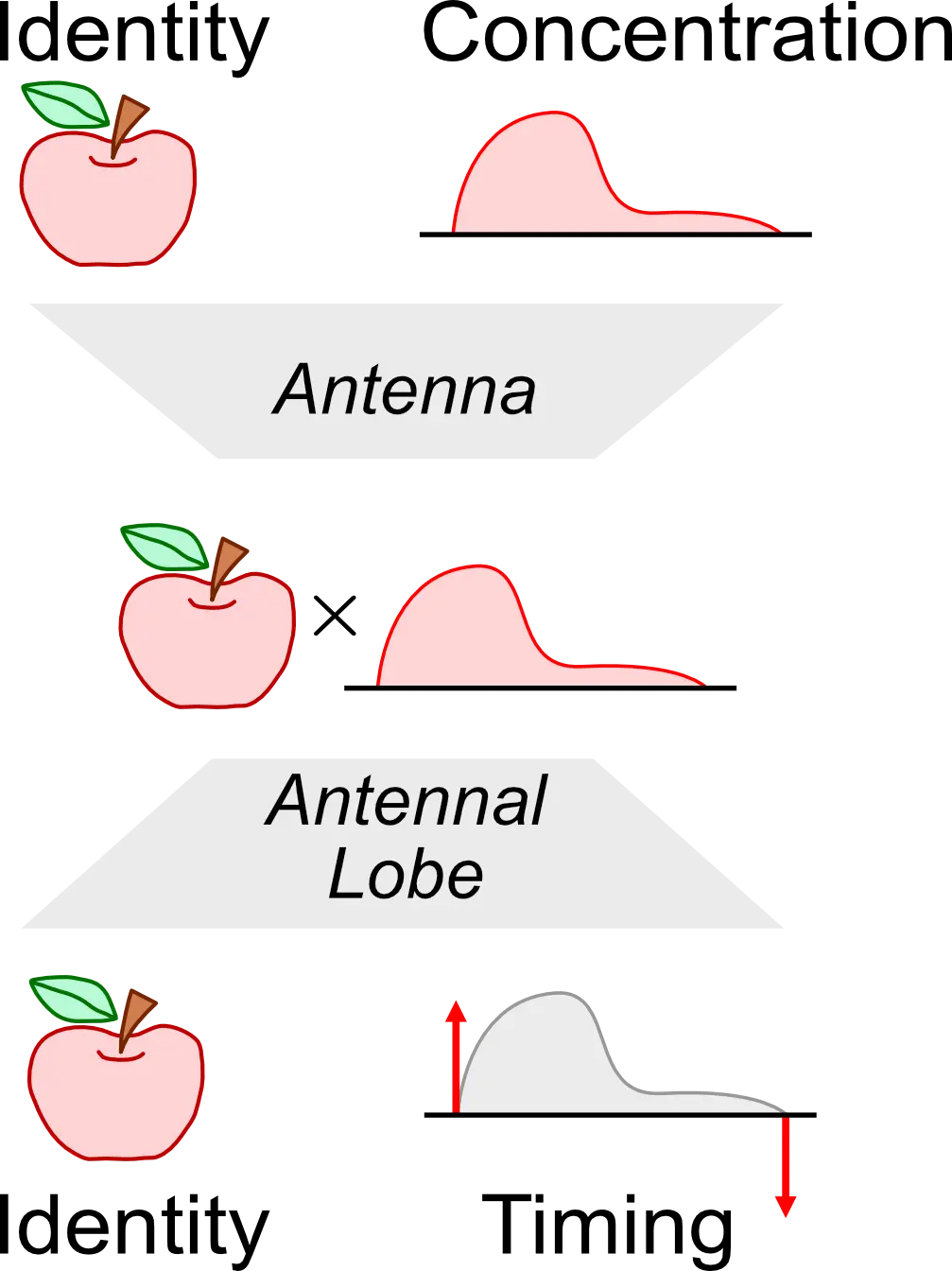 PLOS Computational Biology, Apr 2023
PLOS Computational Biology, Apr 2023We provide theoretical and computational evidence for the functional logic of the Antennal Lobe as a robust odorant object identity recovery processor with ON-OFF event-based processing.
2022
-
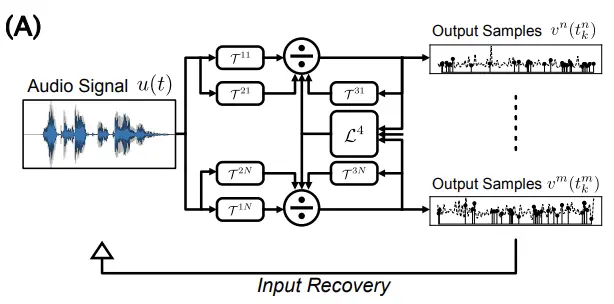 bioRxiv, Sep 2022
bioRxiv, Sep 2022We demonstrate both theoretically and computationally that the Divisive Normalization Processor (DNP) is an invertible operator that can faithfully represent input information given sufficient output samples, with application to different sensory modalities.
-
 bioRxiv, Sep 2022
bioRxiv, Sep 2022We propose a feedback divisive normalization architecture of the Mushroom Body Calyx circuit in the Drosophila olfactory system for odorant demixing. We show that the biological network is highly optimized for processing odorant mixtures.
2021
-
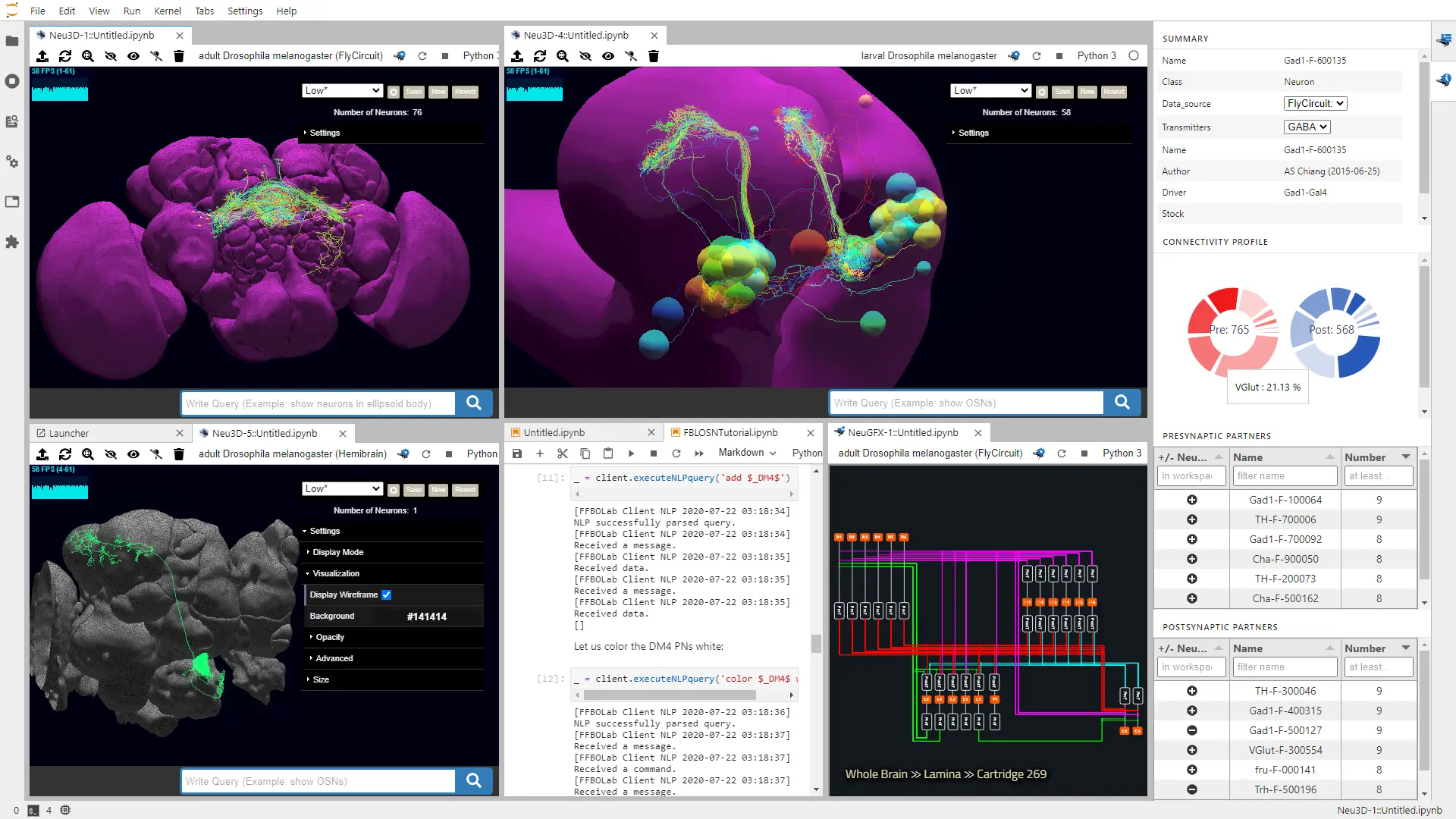 eLife, Feb 2021
eLife, Feb 2021We developed FlyBrainLab, an open-source computing platform that integrates 3D exploration and visualization of diverse datasets with interactive exploration of modeled executable brain circuits in Drosophila.
-
FlyBrainLab: A Complete Programming Environment for Discovering the Functional Logic of the Fruit Fly BrainNeuroinformatics Assembly 2021, Apr 2021
2020
-
 ICASSP 2020, May 2020
ICASSP 2020, May 2020We present a bio-mimetic odorant encoding machine for sampling, reconstruction, and robust representation of odorant identity in the Drosophila olfactory system.
-
FlyBrainLab: An Interactive Computing Platform for Exploring the Drosophila BrainPyData Global, Nov 2020
-
Comparative Study of Fruit Fly Brain Circuit Models with FlyBrainLabneuromatch 3.0, Oct 2020
-
FlyBrainLab: Interactive Computing in the Connectomic/Synaptomic EraJupytercon, Oct 2020
-
Functional Identification of the Odorant Transduction Process of Drosophila Olfactory Sensory NeuronsBMC Neuroscience 2020, 21(Suppl 1):P225, Jul 2020
-
Comparing Drosophila Neural Circuit Models with FlyBrainLabBMC Neuroscience 2020, 21(Suppl 1):P112, Jul 2020
-
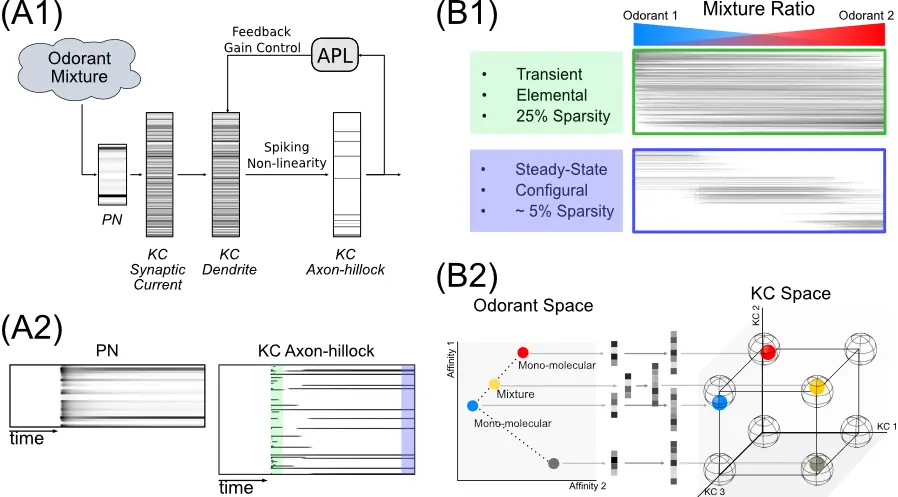 The Geometry of Spatio-Temporal Odorant Mixture Encoding in the Drosophila Mushroom BodyBMC Neuroscience 2020, 21(Suppl 1):218, Jul 2020
The Geometry of Spatio-Temporal Odorant Mixture Encoding in the Drosophila Mushroom BodyBMC Neuroscience 2020, 21(Suppl 1):218, Jul 2020 -
Time-Dependent Simultaneous Configural and Elemental Odorant Mixture ProcessingCOSYNE*2020, Feb 2020
2019
-
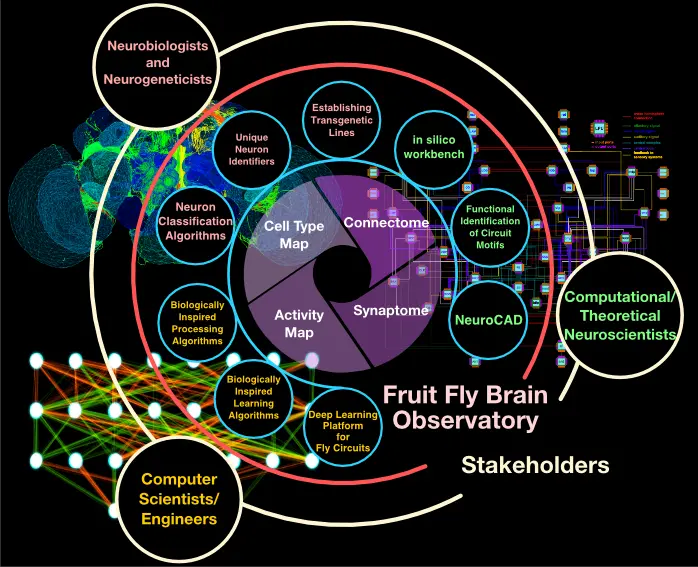 bioRxiv, Mar 2019
bioRxiv, Mar 2019The Fruit Fly Brain Observatory is a platform that aims to bridge the gap between structural and functional data in neuroscience research on Drosophila. It provides tools for exploring and analyzing various types of fruit fly brain data, including connectome and functional imaging data.
-
FlyBrainLab: An Interactive Computing Environment for the Fruit Fly BrainSociety of Neuroscience, Oct 2019
-
Divisive Normalization Circuits Faithfully Represent Visual, Olfactory and Auditory StimuliBMC Neuroscience 2019, 20 (Suppl 1): P353, Oct 2019
2017
-
An Open-Source Model of the Fruit Fly Larval Mushroom BodyNeurobiology of Drosophila, Oct 2017